Introducing hdfsconnector for R
Introduction
Today we’re excited to introduce a new connector for Distributed R that enables R users to read CSV and ORC files from HDFS. One of its main features is the ability to parallelize file loads across multiple machines, drastically improving performance. This post gives an overview of how the connector was designed.
Parallelizing file load
To reduce the time it takes to load a big file the connector splits the loading across different machines in the cluster.
To understand how this works let’s walk through an example. Say we have a 4GB CSV file (test4GB.csv) and a single machine with 4 executors (4 cores). The connector creates as many partitions as executors. In this case each core will load 1GB of the CSV file. When working with ORC files we don’t have that much flexibility because the level of parallelism is already defined (parallelism == number of stripes in the file). In the ORC case we’ll assign different stripes to different executors.
Once we’ve split the file into multiple chunks we assign each chunk to the best executor. For files stored in HDFS we try to minimize data movement in the cluster. This means we’ll assign the chunk to the executor that has the highest number of HDFS blocks locally. In this case executor 1 has the highest number of blocks corresponding to chunk 1 (red), executor 2 has the highest number of blocks for chunk 2 and so on.
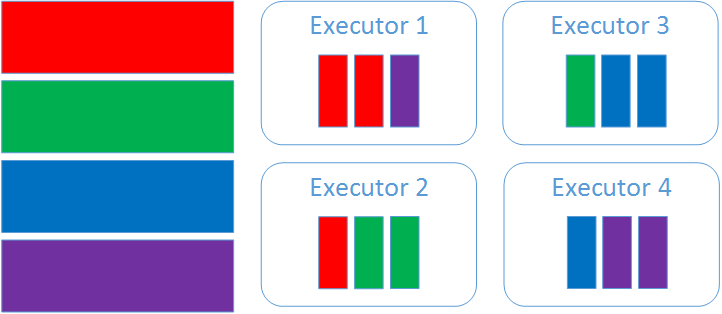
After creating the scheduling map the master sends a message to the executors indicating which chunks should be loaded on which executor. In this case the master sends 1 message to each executor:
- To executor 1: load test4GB.csv, range: [0, 1GB), schema …
- To executor 2: load test4GB.csv, range: [1GB, 2GB), schema …
- To executor 3: load test4GB.csv, range: [2GB, 3GB), schema …
- To executor 4: load test4GB.csv, range: [3GB, 4GB), schema …
Loading individual chunks on the executors
Each executor implements a loading stack made of 4 layers (Assembler, Record Parser, Split Producer and Block Reader). To understand what each layer does it’s useful to start from the bottom. The file is just an array of bytes. We extract splits from the byte array (e.g. lines in CSV). Then we extract individual records from that split (fields in that line). Finally we assemble a final object out of individual records (e.g. an R dataframe).
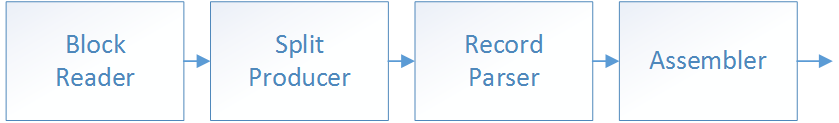
In the next paragraphs we give more details about how the different layers work:
Block reader
Reads a stream of bytes from storage. We have 2 flavors (LocalBlockReader and HdfsBlockReader).
LocalBlockReader reads from the local FS while HdfsBlockReader reads from HDFS. We’re using the WebHDFS interface but we may decide to switch to the native interface in the future if HTTP is not fast enough.
To get the best performance we have an I/O thread that pre-fetches blocks while another thread parses the previous block. The idea is to overlap I/O with compute for improved performance.
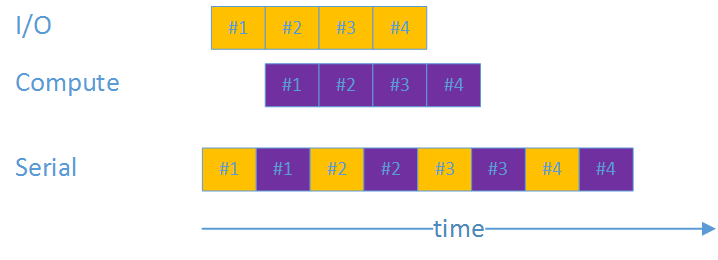
Split producer
Assembles splits (lines in CSV), stripes in ORC, etc. For that it might need to read one or more blocks.
We have 2 flavors DelimiterSplitProducer and OffsetSplitProducer.
In CSV lines are usually determined by a newline character (\n). In this case we use the DelimiterSplitProducer to extract the lines. This is quite slow as we have to look at every byte to detect newline characters. Other formats (such as ORC) include the split boundaries in the metadata and we can extract the splits directly using the OffsetSplitProducer.
An additional complication for CSV files is that the offset from which we start reading may not correspond to the beginning of the line (actually this is the normal case). To handle this we have to backtrack one character and then skip that incomplete line. The discarded line will be processed by the executor that’s working on the previous chunk.
To understand this better let’s use an example. Suppose executor 1 is assigned range [0, 6] and executor 2 range [6,12]. Lines correspond to ranges [0,4),[4,8) and [8,12). Executor 1 will process both lines 1 and 2. Even though the range finishes at offset 6 it will continue until the full 2nd line is consumed. Executor 2 in turn will discard the incomplete line and only process line 3.

Record parser
Breaks a split into individual records. The record parser is format dependent. In the CSV case we use a separator (usually a comma) to separate the records. ORC is a columnar format so all the records in the same column are stored together in a highly optimized way (encoded and compressed).
To understand the importance of the format let’s see how an int column is encoded both in CSV and ORC. Let’s say the data is 1000,1001,1002 … In CSV each record takes 4 bytes or 32 bits (let’s assume the numbers fit between 0 and 9999). In ORC we’d only need 14 bits to encode each number. On top of that we can add smart encodings. For example in this case instead of storing the numbers we can store the delta between each consecutive number. That would be [1,1,1,1 …]. This can be run-length encoded to [num-elements,1]. Finally we can apply compression to reduce the final size even more. ORC supports many different encodings (e.g. dictionary encoding for string type) and different compression types.
Another advantage of ORC vs CSV is that it supports complex types (struct, map, list …)
Assembler
The final layer assembles a number of records into a final object (e.g. an R dataframe).
The mapping in CSV is straightforward. In the ORC case is more complex as it supports more types and they can be arbitrarily nested. The mapping we use is:
- ORC Struct -> R dataframe
- ORC List -> R list
- ORC Map -> R dataframe with 2 columns (key, value).
Performance
Performance is highly dependent on the HW configurations (numbers or cores, performance per core, bisection bandwidth between the HDFS nodes and the nodes loading the files, etc.).
In the graph we present the results of loading a 64MB CSV file with a single machine (4 cores). The time decreases linearly as we add cores.
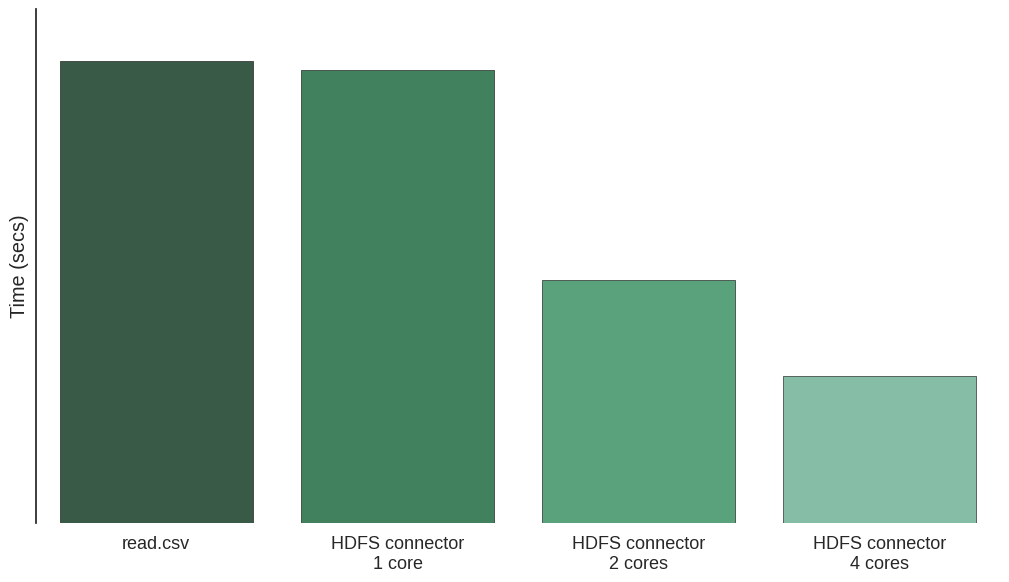
Conclusion
This new connector allows R users to read files from HDFS. It can parallelize file loads across cores and machines which results in great performance.
We offer this functionality as part of Distributed R (supporting distributed operation) and also as an individual package named dataconnector.
The connector is open-source and we encourage feedback and contributions.